
Data Foundations of Precision Immunology
Nov 24, 2025
Graph Team
Now reading:
Data Foundations of Precision Immunology
Author
Graph Team
Date
Nov 24, 2025
11/24/25
Since foundation in 2024, Graph has had the single core aim of building a platform that can scalably and reliably predict what drives immune dysfunction in patients, and do this with biological systems that directly represent the diseased immune systems of patients with autoimmune and inflammatory diseases. This brings clinical reality to early drug discovery in immunology, enabling next-generation medicines for patients with immune-mediated diseases.
Last week at CytoData 2025 in Berlin, we presented work from our rheumatoid arthritis program that demonstrates we can capture disease-specific immune dysfunction at scale with functional readouts combined with multi-omics, and then use that data for prediction of disease biology. It’s very early validation that the approach works, but it’s setting the foundation for a much larger effort at Graph to build a discovery engine bridging dozens of immune-mediated diseases to change how we drive discovery campaigns in I&I for next-generation medicines.
Some background on precision immunology
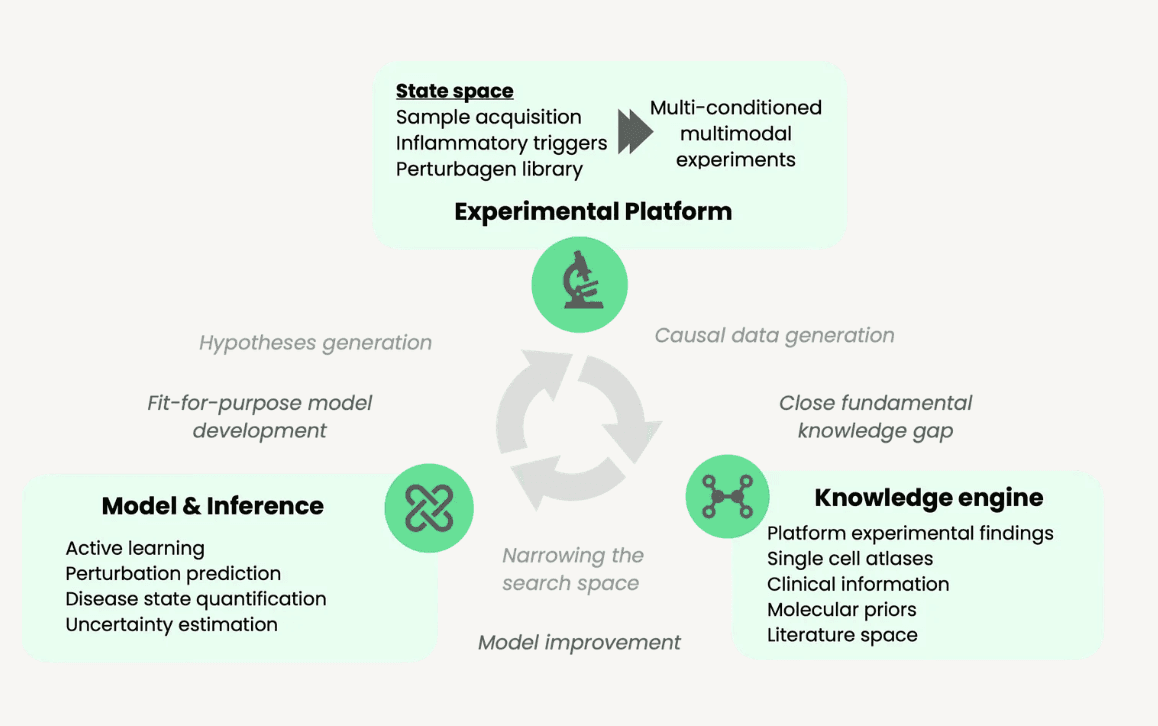
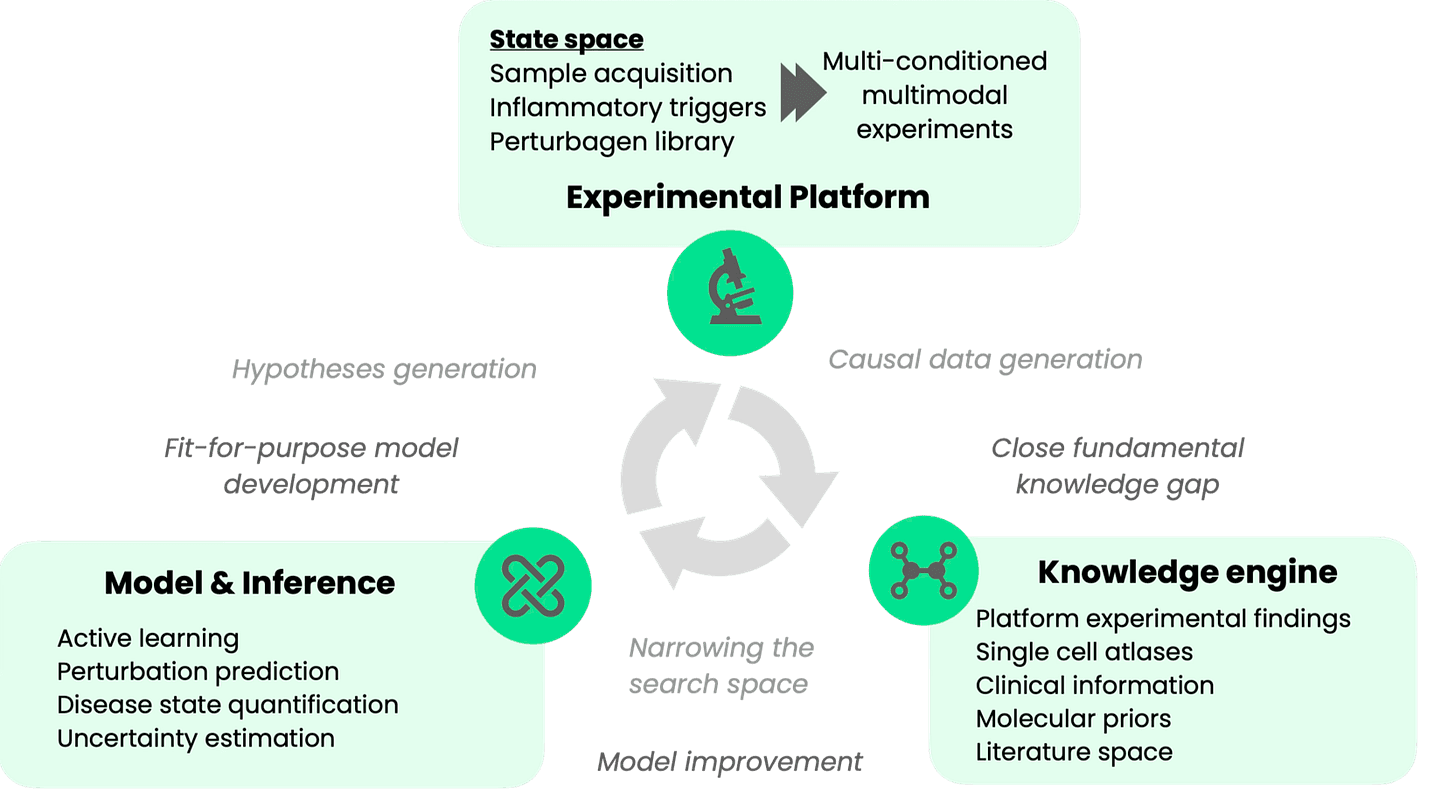
Over the past decade, many of our founding members have been working to close the translational gap between prediction and clinical reality. Graph’s platform combines complex primary patient cell models with lab-in-the-loop AI to systematically test the functional impact of perturbations across diverse disease contexts. At Graph we are building an integrated experimental framework to efficiently distinguish promising from dead-end hypotheses before committing massive drug development investment.
An efficient search strategy is necessary because of the enormous space of immune triggers and contexts, of which current datasets are only scratching the surface. To tackle complex dysregulation, we need to understand immune responses in context, but naive strategies to sample this space quickly run into walls of combinatorial complexity. Graph’s platform maps out the space through smart exploration of live cell responses from donor cohorts, then uses multi-omics to drill down on the biology driving disease-associated changes.
What we've shown at CytoData
In our recent work presented at CytoData we probe cells from rheumatoid arthritis patients to enrich our screen with modulators of an image-based, disease-associated endpoint. In other words, by observing how cells behave when exposed to the disease environment and several hundred known interventions, we build an understanding of what causes immune disease and how to reverse it. We show that the endpoint we identified, an image-based proxy of CD4+ T-cell activation, is a specific biomarker of the dysregulated RA niche - and is modulated by both standard of care drugs and novel predicted drugs.
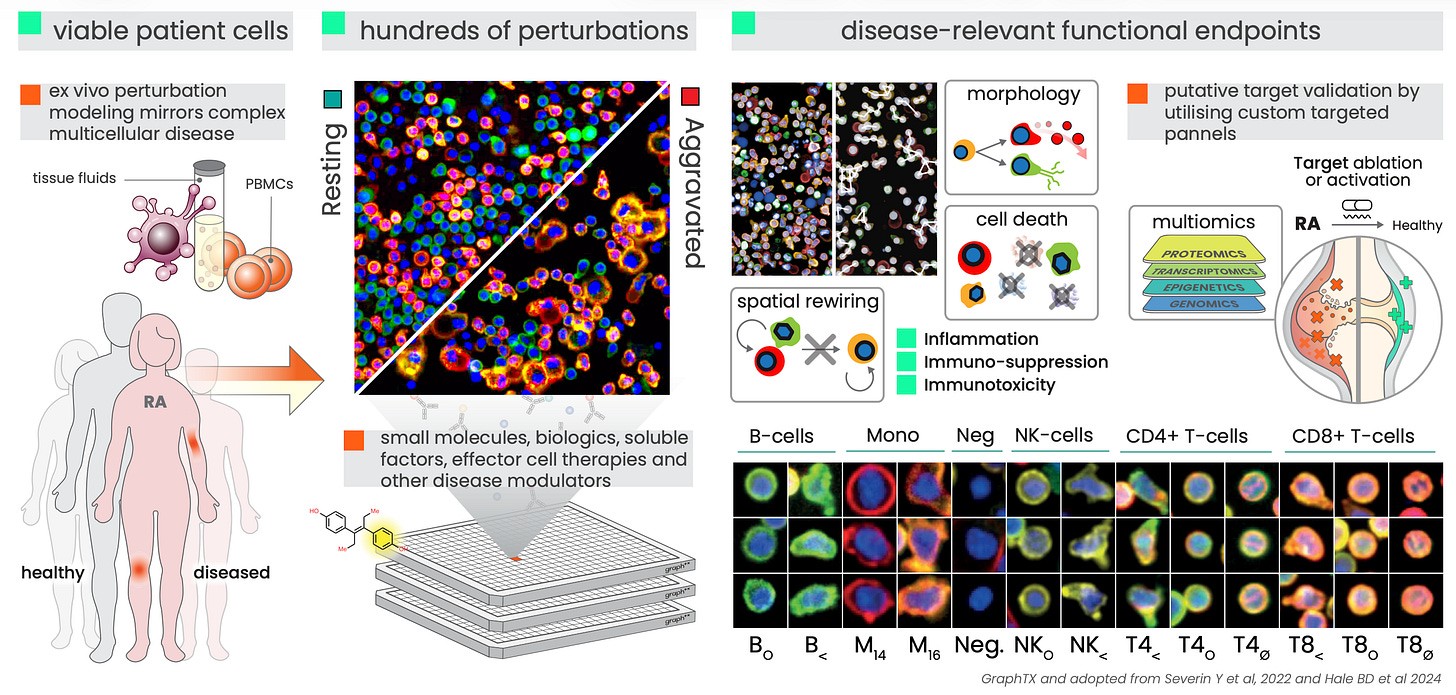
What makes our approach different is that we start by measuring functional associations: highly multiplexed high-content imaging captures cell intrinsic properties, morphological states, and cell-cell interactions, leading us to dynamic functional endpoints that matter for disease and clinical response. When we expose these cells to disease-specific factors and immunomodulators to see how the system responds, pushing cells either deeper into the disease state or pulling them back.
We then went one step further by integrating our screen results with prior knowledge, such as protein-protein interaction networks, to predict new modulators of the disease-associated endpoint. Validating these predictions in the lab, we observed an over 3-fold enrichment over random selection, while bringing us closer to understanding the shared mechanisms of successful modulators. Following up on our functional validation with single cell RNA sequencing and a focused cell migration assay (both before and after perturbation), we were able to connect the observed CD4 activation phenotype to regulation of cell motility.
This story only scratches the surface of what we hope to achieve in future iterations, but gives a first glimpse into the capabilities of our lab-in-the-loop system: catalyzing biological discovery with knowledge integration and AI, while staying grounded in real experimental biology close to the disease niche.
Creating the right data foundation for precision immunology
Despite an explosion of deep (single cell) datasets in recent years, including for immunology, our understanding of context-dependent immune modulation is still too shallow to support faithful virtual cell or even virtual organism models outside of narrow contexts. Approval rates for drug development candidates remain as low as 10%, indicating that a large chunk of development risk is carried all the way to the most expensive stage: clinical trials.
Through a clinically-releavant data-driven approach that starts with patient samples, measures function, incorporates heterogeneity, and validates predictions iteratively, early, we can drastically reduce this risk upfront. Our initial work is a first proof point that the approach works. It sets the stage for functionally guided exploration of the (auto)immune disease space at the much larger scale needed for effective and accurate foundation models of immune biology.
Ultimately, precision immunology requires precision data; not deeper molecular profiling of immortalized cell lines, but dynamic, functional, patient-derived data that actually reflects the complexity of immune-mediated disease context. A key step towards “solving” immunology is to embrace this complexity and strategically map the dynamic repertoire of immune responses.
Graph is building towards context-conditioned foundation models of immune regulation that get smarter with every experiment, creating exponential growth in our understanding of immune disease biology.
We have previously written about why Precision immunology requires precision data and why we think of data as strategic infrastructure. Please find our poster from Cytodata 2025, Buphamalai & Driscoll et al, Decoding the functional and molecular niche of immune-mediated disease for iterative target discovery, here.
Big thanks to the poster and Substack key authors Ize, Katherine, and Robert!
Since foundation in 2024, Graph has had the single core aim of building a platform that can scalably and reliably predict what drives immune dysfunction in patients, and do this with biological systems that directly represent the diseased immune systems of patients with autoimmune and inflammatory diseases. This brings clinical reality to early drug discovery in immunology, enabling next-generation medicines for patients with immune-mediated diseases.
Last week at CytoData 2025 in Berlin, we presented work from our rheumatoid arthritis program that demonstrates we can capture disease-specific immune dysfunction at scale with functional readouts combined with multi-omics, and then use that data for prediction of disease biology. It’s very early validation that the approach works, but it’s setting the foundation for a much larger effort at Graph to build a discovery engine bridging dozens of immune-mediated diseases to change how we drive discovery campaigns in I&I for next-generation medicines.
Some background on precision immunology
Over the past decade, many of our founding members have been working to close the translational gap between prediction and clinical reality. Graph’s platform combines complex primary patient cell models with lab-in-the-loop AI to systematically test the functional impact of perturbations across diverse disease contexts. At Graph we are building an integrated experimental framework to efficiently distinguish promising from dead-end hypotheses before committing massive drug development investment.
An efficient search strategy is necessary because of the enormous space of immune triggers and contexts, of which current datasets are only scratching the surface. To tackle complex dysregulation, we need to understand immune responses in context, but naive strategies to sample this space quickly run into walls of combinatorial complexity. Graph’s platform maps out the space through smart exploration of live cell responses from donor cohorts, then uses multi-omics to drill down on the biology driving disease-associated changes.
What we've shown at CytoData
In our recent work presented at CytoData we probe cells from rheumatoid arthritis patients to enrich our screen with modulators of an image-based, disease-associated endpoint. In other words, by observing how cells behave when exposed to the disease environment and several hundred known interventions, we build an understanding of what causes immune disease and how to reverse it. We show that the endpoint we identified, an image-based proxy of CD4+ T-cell activation, is a specific biomarker of the dysregulated RA niche - and is modulated by both standard of care drugs and novel predicted drugs.

What makes our approach different is that we start by measuring functional associations: highly multiplexed high-content imaging captures cell intrinsic properties, morphological states, and cell-cell interactions, leading us to dynamic functional endpoints that matter for disease and clinical response. When we expose these cells to disease-specific factors and immunomodulators to see how the system responds, pushing cells either deeper into the disease state or pulling them back.
We then went one step further by integrating our screen results with prior knowledge, such as protein-protein interaction networks, to predict new modulators of the disease-associated endpoint. Validating these predictions in the lab, we observed an over 3-fold enrichment over random selection, while bringing us closer to understanding the shared mechanisms of successful modulators. Following up on our functional validation with single cell RNA sequencing and a focused cell migration assay (both before and after perturbation), we were able to connect the observed CD4 activation phenotype to regulation of cell motility.
This story only scratches the surface of what we hope to achieve in future iterations, but gives a first glimpse into the capabilities of our lab-in-the-loop system: catalyzing biological discovery with knowledge integration and AI, while staying grounded in real experimental biology close to the disease niche.
Creating the right data foundation for precision immunology
Despite an explosion of deep (single cell) datasets in recent years, including for immunology, our understanding of context-dependent immune modulation is still too shallow to support faithful virtual cell or even virtual organism models outside of narrow contexts. Approval rates for drug development candidates remain as low as 10%, indicating that a large chunk of development risk is carried all the way to the most expensive stage: clinical trials.
Through a clinically-releavant data-driven approach that starts with patient samples, measures function, incorporates heterogeneity, and validates predictions iteratively, early, we can drastically reduce this risk upfront. Our initial work is a first proof point that the approach works. It sets the stage for functionally guided exploration of the (auto)immune disease space at the much larger scale needed for effective and accurate foundation models of immune biology.
Ultimately, precision immunology requires precision data; not deeper molecular profiling of immortalized cell lines, but dynamic, functional, patient-derived data that actually reflects the complexity of immune-mediated disease context. A key step towards “solving” immunology is to embrace this complexity and strategically map the dynamic repertoire of immune responses.
Graph is building towards context-conditioned foundation models of immune regulation that get smarter with every experiment, creating exponential growth in our understanding of immune disease biology.

We have previously written about why Precision immunology requires precision data and why we think of data as strategic infrastructure. Please find our poster from Cytodata 2025, Buphamalai & Driscoll et al, Decoding the functional and molecular niche of immune-mediated disease for iterative target discovery, here.
Big thanks to the poster and Substack key authors Ize, Katherine, and Robert!
Read More
Graph partners with Parse Biosciences
Graph partners with Parse Biosciences
News
Jan 20, 2026
1/20/26
Data Foundations of Precision Immunology
Data Foundations of Precision Immunology
Article
Nov 24, 2025
11/24/25
First data in RA - Cytodata 2025
First data in RA - Cytodata 2025
Research
Dec 11, 2025
12/11/25
Graph and BIIE announce strategic collaboration to in precision immunology
Graph and BIIE announce strategic collaboration to in precision immunology
News
Oct 7, 2025
10/7/25
Graph awarded €1.1M Deep Tech Grant
Graph awarded €1.1M Deep Tech Grant
News
Apr 22, 2025
4/22/25
GTX co-founder appointed as Faculty Professor at the BIIE
GTX co-founder appointed as Faculty Professor at the BIIE
News
Mar 28, 2025
3/28/25
Graph Awarded €1.1M "Austrian Life Sciences" FFG Grant
Graph Awarded €1.1M "Austrian Life Sciences" FFG Grant
News
Mar 3, 2025
3/3/25
Graph Raises $3.1M Pre-Seed Round
Graph Raises $3.1M Pre-Seed Round
News
Dec 10, 2025
12/10/25
Copyright GraphTx 2026
Vienna, Asutria
Vienna, Austria
All system opperational